The global problem of COVID-19
Coronavirus disease 2019 (COVID-19) pandemic has been a widespread global public health issue, with 90 602 820 reported confirmed cases and 1939 959 confirmed deaths in 216 countries, areas or territories by 11 January 2021, it infected more population than SARS with 8273 diagnosed cases and 775 deaths and MERS with 1139 diagnosed cases and 431 deaths caused in 2003 and 2013, respectively [1,2]. Due to lacking of potent therapeutic ways to treat this disease, even the countries with the good healthcare facilities are reeling under the burden of COVID-19. Meanwhile, low- and middle-income countries (LMICs) have been severely hit by the outbreak of COVID-19 and face the fact of infrastructure lacking and problem of overcrowding, making it even more challenging to limit the spread of COVID-19 [3].
The SARS-Cov-2 virus
SARS-CoV-2 was isolated from airway epithelial cells of COVID-19 patients on January 12, 2020. Phylogenetic analysis of full-length genome sequences of SARS-CoV-2 showed that the genome sequence of SARS-CoV-2 is highly similar to SARS-CoV (about 79%), and SARS-CoV-2 uses the same cell entry receptor, ACE2, as SARS-CoV does [4,5], however, SARS-CoV-2 binds to ACE2 with an even higher affinity than SARS-CoV, which could partly explain why SARS-CoV-2 is much more infectious than SARS-CoV. Moreover, the spike protein of SARS-CoV-2 has a lower free energy than that of SARS-CoV, indicating that SARS-CoV-2 may survive under a higher temperature than SARS-CoV [6].
The SARS-Cov-2 proteins
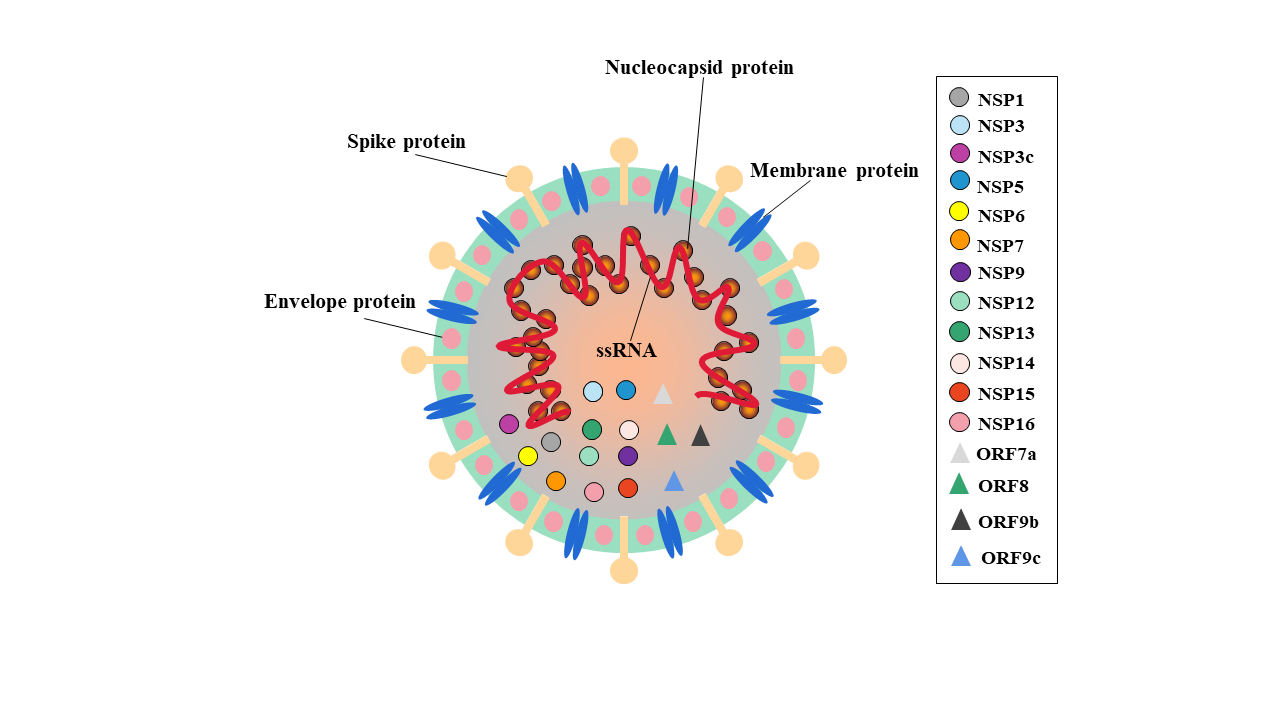
SARS-CoV-2 is an ssRNA virus and its genome is 29903nt in length. There are six functional open reading frames (ORFs) arranged in order from 5’ to 3’: ORF1a/ORF1b (replicase), ORF2 (Spike protein, S), ORF4 (Envelope protein, E), ORF5 (Membrane protein, M), ORF9 (Nucleocapsid protein, N). Moreover, several putative ORFs encode accessory proteins. The S1 subunit of S protein promotes ACE2 mediated virus attachment while S2 subunit enhances membrane fusion. Besides, the N protein is formed of a serine-rich linker region sandwiched between N Terminal Domain (NTD) and C Terminal Domain (CTD), plays a crucial role in viral entry and its processing post entry. The E protein contains a NTD, hydrophobic domain and CTD which form viroporins needed for viral assembly. The M protein has hydrophilic C terminal and amphipathic N terminal, tts long-form facilitates spike incorporation and the interaction with E promotes virion production [7]. The late entry of vaccines and limited effective treatments for the aggressive COVID-19 has made a sense of urgency for the discovery of potent drugs. Therefore, the discovery or design of drugs that can bind to the virus proteins and inhibit its function is a potential way to combat the virus.
dbSCI: Main features
Considering the great efforts for reducing the COVID-19 burden worldwide, numbers of researches are approaching to explore the molecular infection mechanisms, virus evolutionary aspects and drug development of SARS-CoV-2. Within this study, we developed database naming as dbSCI ( database of SARS-CoV-2 Inhibitors for COVID-19 ). dbSCI includes 234 SARS-CoV-2 inhibitors collected from publications based on cell experiments, 86 drugs of COVID-19 in clinical trials and 1305 potential SARS-CoV-2 inhibitors from bioinformatics analysis. 2777 manually curated entries were obtained, and each entry encompasses comprehensive information regarding of inhibitor/drug name, targeting protein, functional mechanism of inhibitor, experimental technique, experimental sample type (cell line and/or tissue), reference information (PubMed ID, year of publication, title of paper).
References
1. World Health Organization. Coronavirus disease (COVID-19) pandemic. Available at https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed: 19 September 2020.
2. Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92(6):568-576. doi:10.1002/jmv.25748.
3. Angrup A, Kanaujia R, Ray P, Biswal M. Healthcare facilities in low- and middle-income countries affected by COVID-19: Time to upgrade basic infection control and prevention practices. Indian J Med Microbiol. 2020;38(2):139-143. doi:10.4103/ijmm.IJMM_20_125.
4. Wang H, Li X, Li T, et al. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol Infect Dis. 2020;39(9):1629-1635. doi:10.1007/s10096-020-03899-4.
5. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565-574. doi:10.1016/S0140-6736(20)30251-8.
6. He J, Tao H, Yan Y, Huang SY, Xiao Y. Molecular Mechanism of Evolution and Human Infection with SARS-CoV-2. Viruses. 2020;12(4):428. Published 2020 Apr 10. doi:10.3390/v12040428.
7. Satarker S, Nampoothiri M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch Med Res. 2020;51(6):482-491. doi:10.1016/j.arcmed.2020.05.012.