| OStme is a web server for analyzing the TME components of 23917 tumor samples with clinical survival information from the TCGA, GEO, CGGA database and literature, using four algorithms including CIBERSORTx, xCell, EPIC and ESTIMATE.
Statement: Use the Internet Explorer or Google Chrome browser for best visualization quality. |
2.1 Home
Four types of functional analyses are supported in OStme (Figure 1): 'Survival analysis' refers to survival analysis of given TIC in a selected cancer type; 'Differencial analysis' refers to the fraction variations of selected TIC across different stages/grades or variations regarding therapies by using different algorithms; 'Infiltration estimation' refers to the visual fraction of all TICs across different tumors. 'Infiltration association' refers to the relationship between gene of interested and all TICs across different tumors. The material of Home figure was provided by Servier Medical Art (https://smart.servier.com) under CC BY 3.0 license.


Figure 1. Four quick functional module in home pages.
2.2 Survival analysis
This feature “Survival analysis” refers to survival analysis of given immune cell type(s) in a selected cancer type (Figure 2). OStme performs 13 survival items including OS, DFS based on cell fractions of TME. OStme uses Log-rank p-value for hypothesis test. The cox proportional hazard ratio and the 95% confidence interval information can also be included in the survival plot. Cell types most related with patient survival can be searched. Users can choose interested cell type, computational tool, one cancer type, dataset source, survival items, group split, stage, gender. Then the KM plot results will be shown.
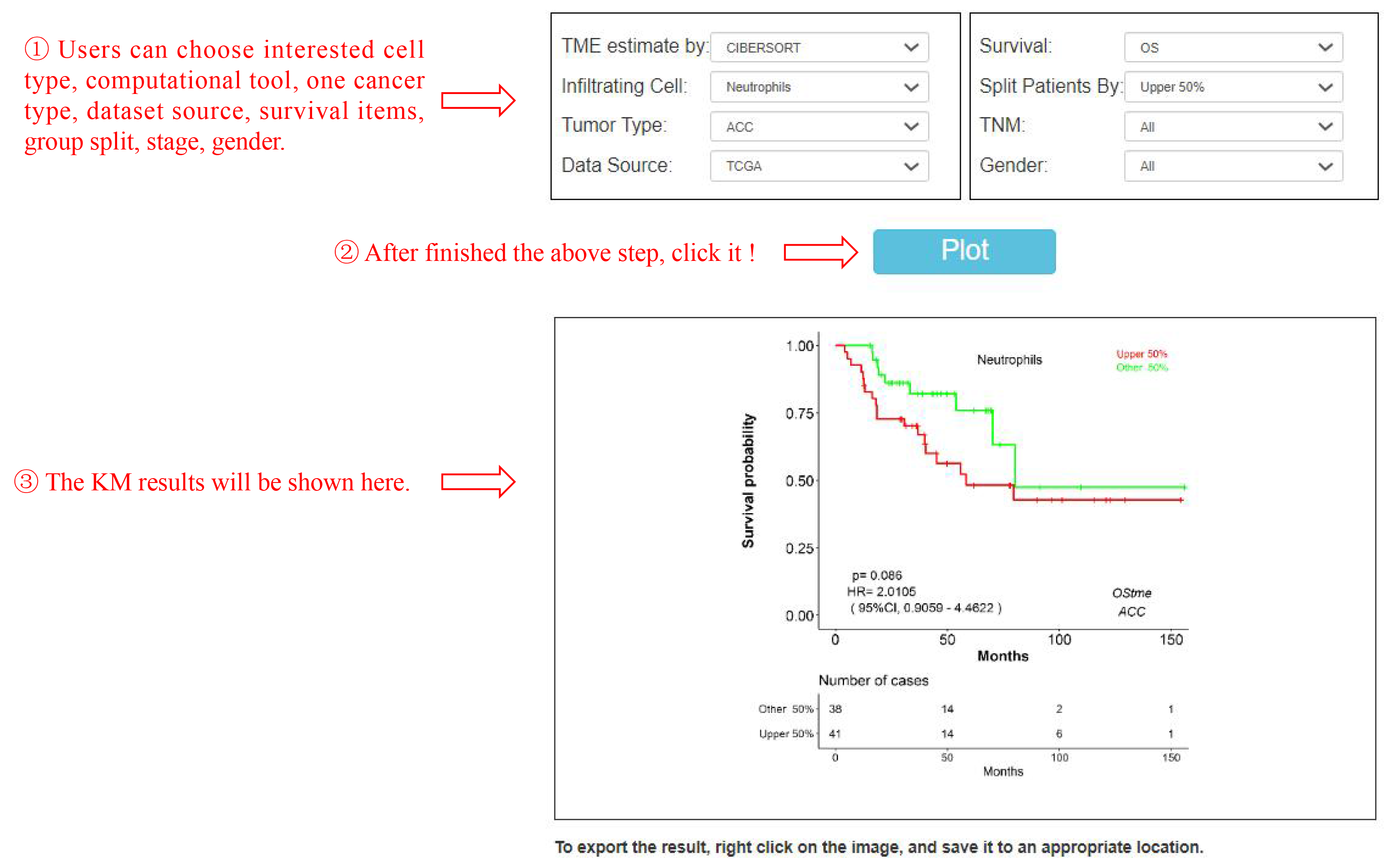 Figure 2. ACC patients with high abundance of neutrophils shows poor overall survival (OS) than those patients with low abundance of neutrophils. Red line represents upper 50% of ACC patients with high abundance of neutrophils. Green line represents other 50% of ACC patients with low abundance of neutrophils.
Figure 2. ACC patients with high abundance of neutrophils shows poor overall survival (OS) than those patients with low abundance of neutrophils. Red line represents upper 50% of ACC patients with high abundance of neutrophils. Green line represents other 50% of ACC patients with low abundance of neutrophils.
 Figure 2. ACC patients with high abundance of neutrophils shows poor overall survival (OS) than those patients with low abundance of neutrophils. Red line represents upper 50% of ACC patients with high abundance of neutrophils. Green line represents other 50% of ACC patients with low abundance of neutrophils.
Figure 2. ACC patients with high abundance of neutrophils shows poor overall survival (OS) than those patients with low abundance of neutrophils. Red line represents upper 50% of ACC patients with high abundance of neutrophils. Green line represents other 50% of ACC patients with low abundance of neutrophils.
2.3 Differential analysis
This feature "Differential analysis" generates expression violin plots based on patient stage, grades and treatment response(Figure 3). Here users can choose an interested clinical characteristics, computational tool, cell type, tumor type, and dataset source. Click the “Plot” button: OStme will generate a TME cell fractions plot based on user custom inputs.
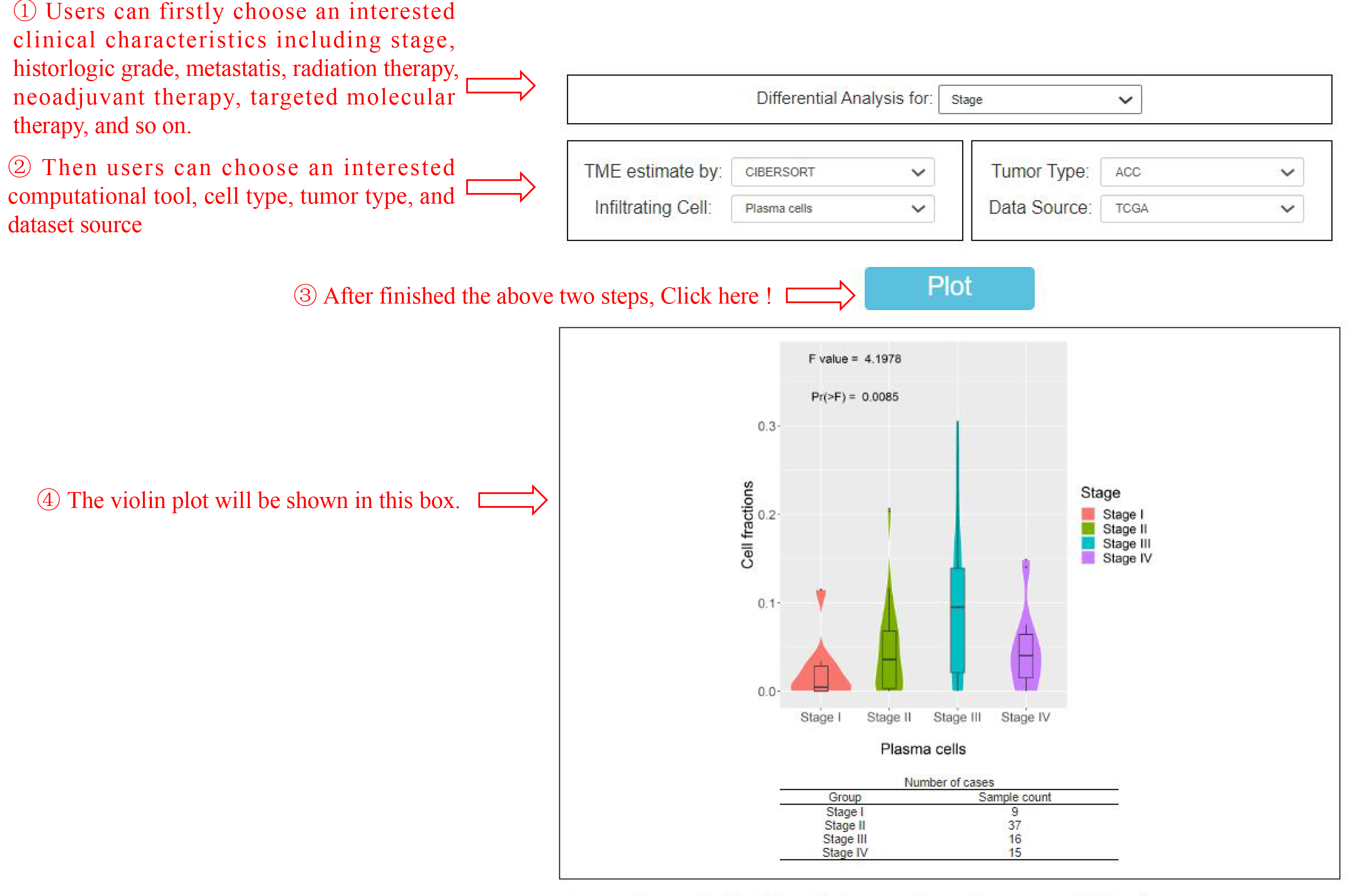 Figure 3. The abundance of plasma cells in different stage of ACC patients from TCGA dataset.
Figure 3. The abundance of plasma cells in different stage of ACC patients from TCGA dataset.
 Figure 3. The abundance of plasma cells in different stage of ACC patients from TCGA dataset.
Figure 3. The abundance of plasma cells in different stage of ACC patients from TCGA dataset.
2.4 Infiltration estimation
This feature "Infiltration estimation" provides fractions matrix heatmap based on computer tool and tumor type (Figure 4).
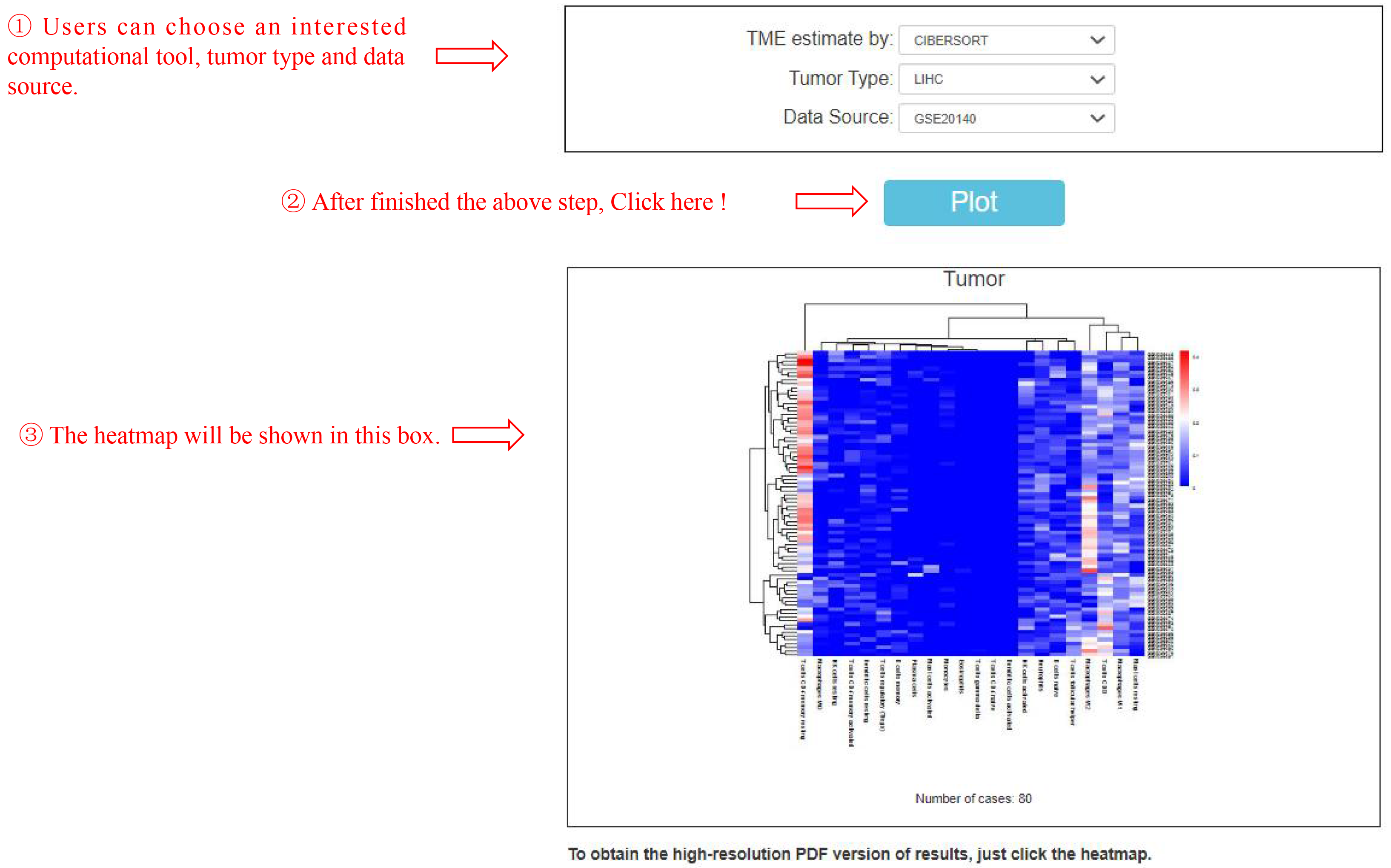 Figure 4. Heatmap shows the relative abundance of 22 types of infiltration cell in LIHC from TCGA dataset. The density of color in each block represents the cell fraction value of tumor tissue.
Figure 4. Heatmap shows the relative abundance of 22 types of infiltration cell in LIHC from TCGA dataset. The density of color in each block represents the cell fraction value of tumor tissue.
 Figure 4. Heatmap shows the relative abundance of 22 types of infiltration cell in LIHC from TCGA dataset. The density of color in each block represents the cell fraction value of tumor tissue.
Figure 4. Heatmap shows the relative abundance of 22 types of infiltration cell in LIHC from TCGA dataset. The density of color in each block represents the cell fraction value of tumor tissue.
2.5 Infiltration association
This feature "Infiltration association" refers to analyze the correlation between expression of interested gene and the abundance of infiltration cell in various cancer types (Figure 5).
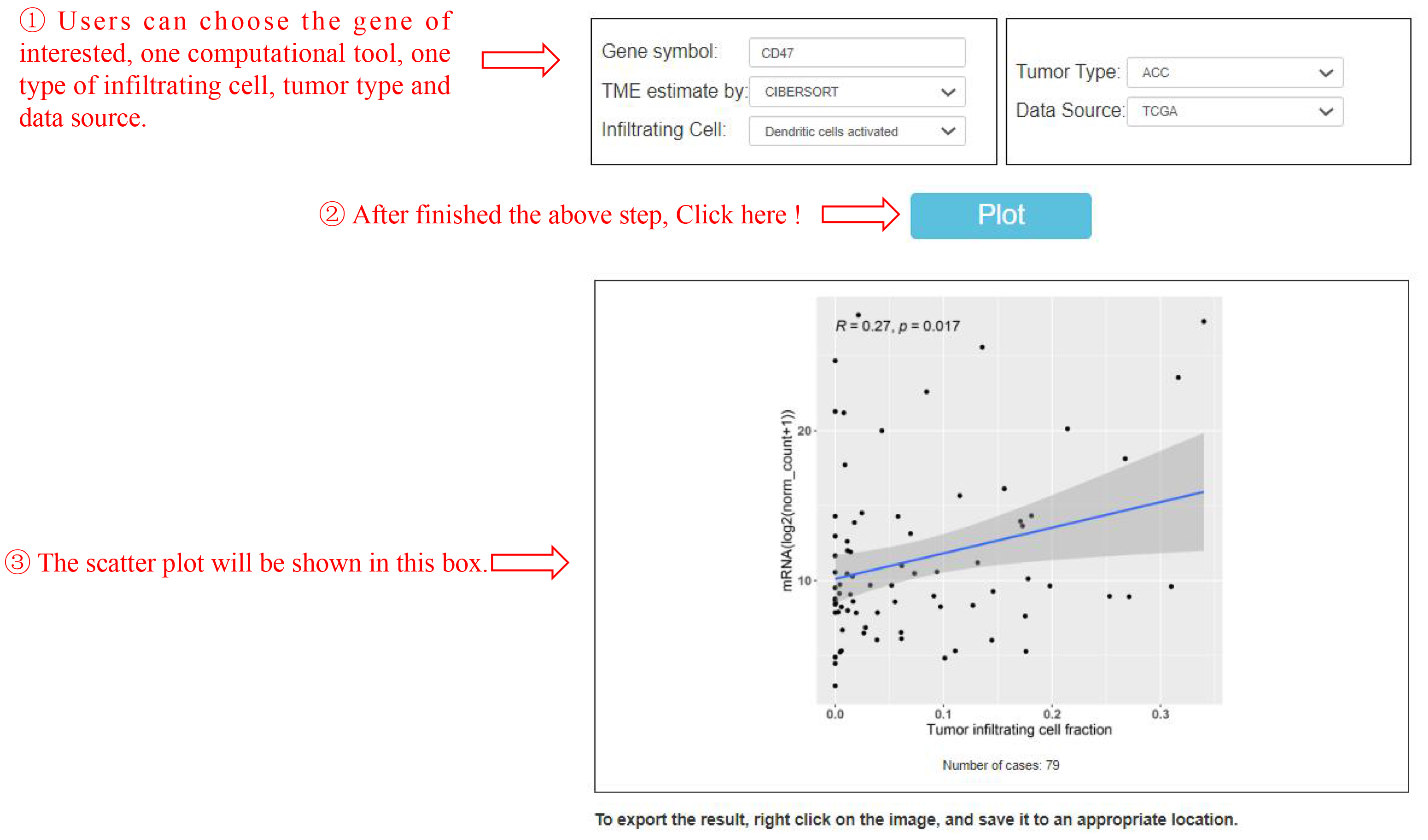 Figure 5. The association between CD47 gene expression and the abundance of dendritic cells activated in ACC. The strength can be evaluated by these general guidelines (which may vary by discipline): 0.1<|R|<0.3 represents small/weak correlation; 0.3<|R|<0.5 represents medium/moderate correlation; |R|>0.5 large/strong correlation.
Figure 5. The association between CD47 gene expression and the abundance of dendritic cells activated in ACC. The strength can be evaluated by these general guidelines (which may vary by discipline): 0.1<|R|<0.3 represents small/weak correlation; 0.3<|R|<0.5 represents medium/moderate correlation; |R|>0.5 large/strong correlation.
 Figure 5. The association between CD47 gene expression and the abundance of dendritic cells activated in ACC. The strength can be evaluated by these general guidelines (which may vary by discipline): 0.1<|R|<0.3 represents small/weak correlation; 0.3<|R|<0.5 represents medium/moderate correlation; |R|>0.5 large/strong correlation.
Figure 5. The association between CD47 gene expression and the abundance of dendritic cells activated in ACC. The strength can be evaluated by these general guidelines (which may vary by discipline): 0.1<|R|<0.3 represents small/weak correlation; 0.3<|R|<0.5 represents medium/moderate correlation; |R|>0.5 large/strong correlation.
| Xie et al. OStme: Online consensus Survival analysis webserver of Tumor MicroEnvironment components of pan-cancers. 2022 |
