


| dbTAMP is a powerful database that could help researchers to explore mechanisms of tumor-associated macrophages polarization. The database of dbTAMP, which covers 668 genes (527 protein-encoding genes and 141 protein non-coding genes) and 150 drugs related with TAMs derived from 13857 published articles in PubMed. The number of M1 markers is 20, while there are 16 M2 markers simultaneously. It provides not only a user-friendly interface for searching, browsing but also four analysis modules for survival analysis, differential analysis, infiltration estimation and infiltration association. |

|
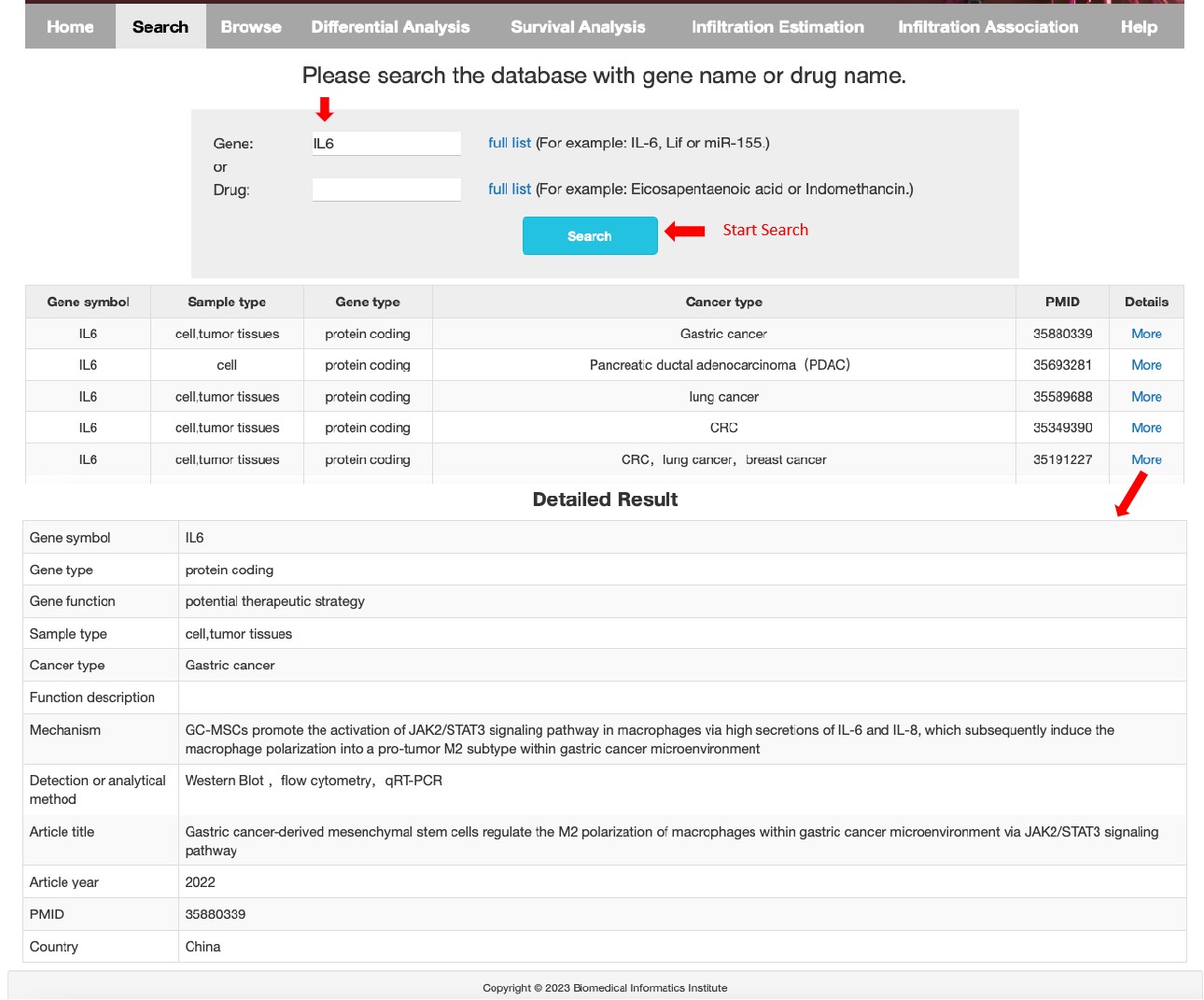
Search:
dbTAMP provides 2 query search methods for users to obtain the data, including ‘Search by gene’ (select and input gene symbols of interest), ‘Search by drug’ (select a drug name of interest) . Input the gene or drug of interest and some key information is revealed. (Gene symbol, sample type, Gene type, Cancer type and so on). Clicking on “More” allows user to download the browsing table.
Browse:
The browse page provides users with four options: browse by Protein-coding genes, Non-coding genes, Drugs or Marker. On the browse page, it contains 6 categories: Gene /Drug symbol, sample type, Gene type, Cancer type, Function description, and PMID. Clicking on “More” allows user to download the browsing table.


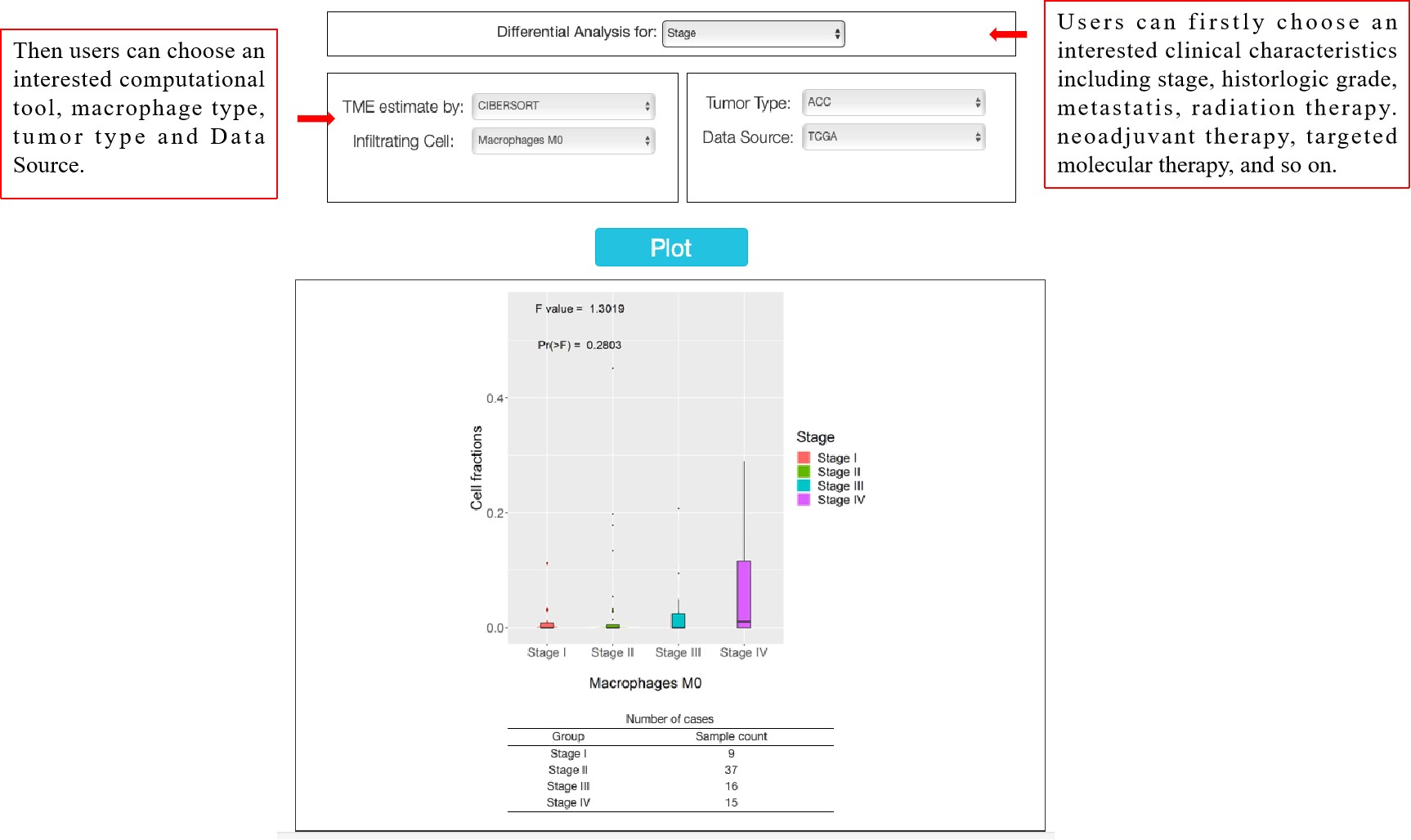
Differential Analysis:
Two datasets were included, differential genes for M1 and M2 in tumors, and differences in macrophage composition in normal and tumor tissues. This feature "Differential analysis" generates expression violin plots based on patient stage, grades and treatment response. Here users can choose an interested clinical characteristics, computational tool, macrophage type, tumor type, and dataset source. Click the “Plot” button: dbTAMP will generate a macrophage fractions plot based on user custom inputs.

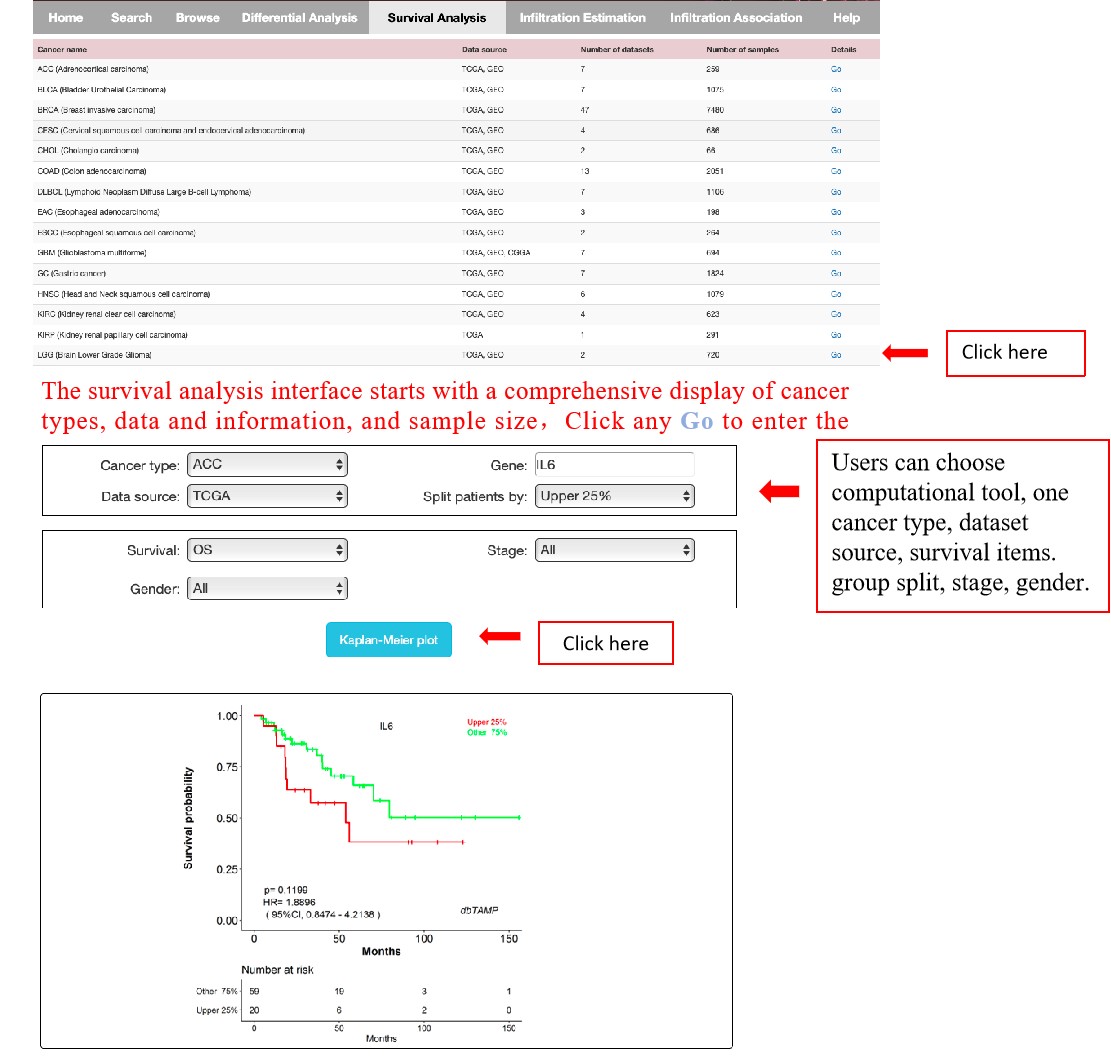
Survival analysis:
This feature “Survival analysis” refers to survival analysis of given macrophage in a selected cancer type. dbTAMP performs 13 survival items including OS, DFS based on cell fractions of macrophage. dbTAMP uses Log-rank p-value for hypothesis test. The cox proportional hazard ratio and the 95% confidence interval information can also be included in the survival plot. Users can choose computational tool, one cancer type, dataset source, survival items, group split, stage, gender. Then the KM plot results will be shown.

Infiltration estimation:
This feature "Infiltration estimation" provides fractions matrix heatmap based on computer tool and tumor type .

Heatmap shows the relative abundance of 22 types of infiltration cell in ACC from TCGA dataset. The density of color in each block represents the cell fraction value of tumor tissue.

Heatmap shows the relative abundance of 22 types of infiltration cell in ACC from TCGA dataset. The density of color in each block represents the cell fraction value of tumor tissue.
Infiltration association:
This feature "Infiltration association" refers to analyze the correlation between expression of interested gene and the abundance of macrophage type in various cancer types.

The association between LRRC32 gene expression and the abundance of M2 type of macrophage activated in STAD. The strength can be evaluated by these general guidelines (which may vary by discipline): 0.1<|R|<0.3 represents small/weak correlation; 0.3<|R|<0.5 represents medium/moderate correlation; |R|>0.5 large/strong correlation.

The association between LRRC32 gene expression and the abundance of M2 type of macrophage activated in STAD. The strength can be evaluated by these general guidelines (which may vary by discipline): 0.1<|R|<0.3 represents small/weak correlation; 0.3<|R|<0.5 represents medium/moderate correlation; |R|>0.5 large/strong correlation.
| The articles in the database and genetic screening are sourced exclusively from the pubmed website. The data used in dbTAMP come from TCGA (The Cancer Genome Atlas) and GEO (Gene Expression Omnibus). The typies of data used in dbTAMP are RNA seq (TCGA) and Gene chip (GEO). |
Copyright © 2023 Biomedical Informatics Institute