Search:
In “Search” page, users can just input a sample ID or an interesting gene/drug name to obtain the detailed information of a certain gene/drug of CAC.
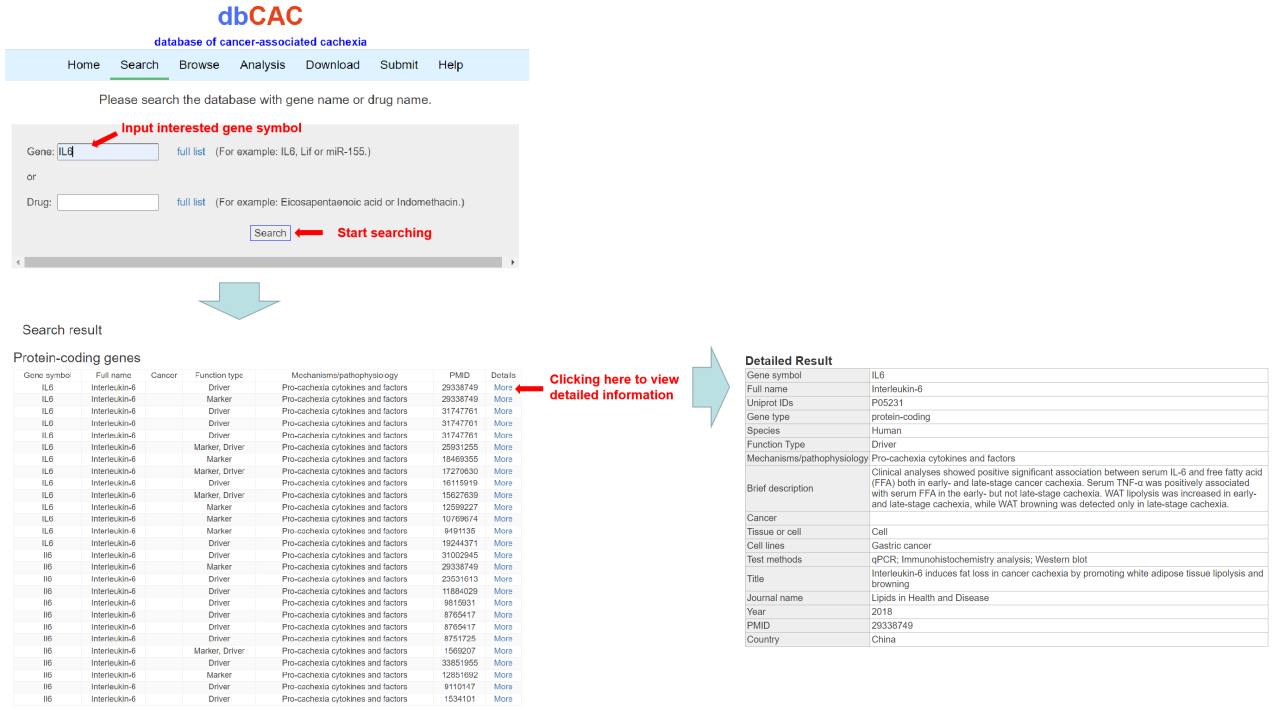
1. Search by gene symbol or protein name.
If users want to query a gene or protein by name, e.g. IL6, users can input “IL6” in the box and press “Search” button. The results will list all potential entries related to inputted “IL6”. The detailed information about the gene and CAC is obtained by clicking “More”.
2. Search by drug name.
Example: If users want to query a potential drug, which can inhibit CAC, e.g. acadesine, users should input the drug name “acadesine” in the drug name box. The results list all existing entries related to usersr inputted “acadesine”. The detailed information about the drug and CAC are obtained by clicking “More”.

Browse:
This module includes 3 parts such as “Protein-coding genes”, “non-coding genes”, “Drugs”. Users can browse any parts autonomously, which users are interested in, e.g Protein coding genes. Then, click on “Protein coding genes” and all selected entries will be presented.

Analysis:
We also provide the analysis function for protein-encoding genes to predict their prognostic value in multiple cancer types. If users want to query a gene or protein by gene symbol, e.g. IL6, users can input “IL6” in the box and press “Analysis” button. The table list containing HR, 95%CI, P value of IL6 in different gene expression datasets of various cancers is shown. If users want to view the survival curve, users can click the “Go”. Then users can choose the cut off value (e.g upper 25%, upper 30%, upper 50% and so on), and click the “Kaplan-Meier plot”. Finally, the results of KM plot will be shown.
